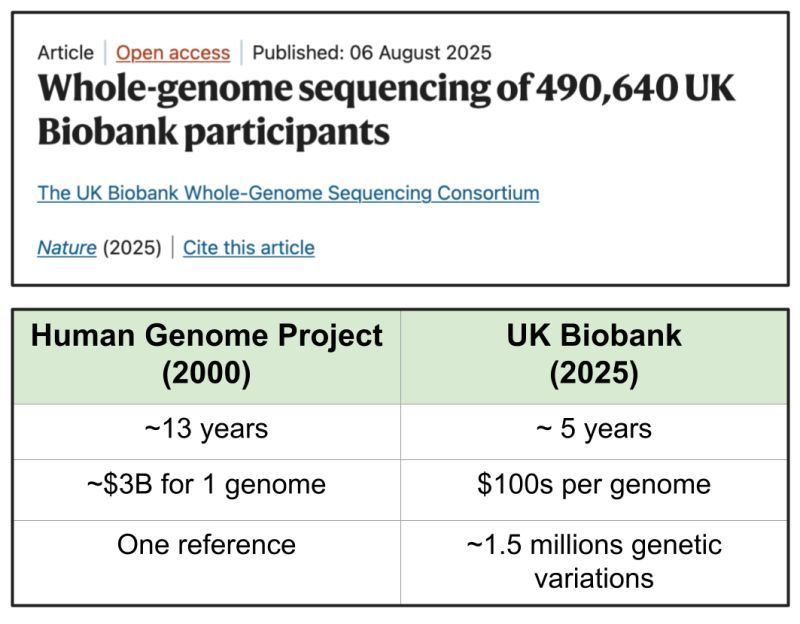
To put this into context, the Human Genome Project built a single human genome reference by sequencing DNA from a handful of people. It cost ~$3B and took 13 years. The latest work is nearly half a million people, done at a rate of ~115,000 genomes/year, and a few hundred dollars per genome.
Replies
A big challenge in genetics is linking genetic variations to an outcome (disease). Variations can be single DNA base changes, additions or deletions of DNA, which are found in a percentage of the population In this latest paper, they identified ~1.5 billion variations.
And each UK biobank participant provides extensive phenotypic data (eg health questionnaires, demographics, proteomics, metabolomics…). So this is a major step in linking genetic variation to diseases and improving diagnostics.
retrospectively
~1 billion SNPs in a ~3 billion base genome, so with 4 bases that is 250M SNP positions (assuming no bias for coding or regulatory regions) or roughly 1/12 bases in our genome has a possible variant between individuals. With a sample size of 500k genomes, is it even possible to find this many SNPs?
A large number of unique SNPs - eg in only 1 individual could be sequencing errors - cause no way they have enough sample size of sequencing coverage to say for sure. But in the end they have 1,359 significant gene–phenotype associations to play with, which is great.