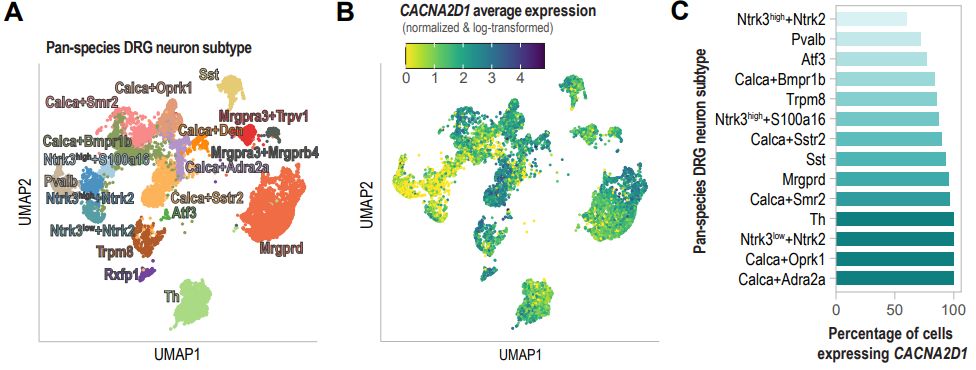
In this case I wonder whether (C) communicated this idea more clearly and succinctly, and could have been presented on its own or with other types of data. What the UMAP is doing is showing the large dataset you had & how diverse DRG are, but it could be misleading re: how similar/different they are
Replies
I think what I got from the episode, is it's ok to use for trying to understand if 'clusters' exist, but the bottom line is real data needs to back up the UMAP. Additionally, be careful about what kind of data you use. RNA-seq may be a good use bc it exists on a manifold from low to high expression.
That’s a good point about RNAseq! The problem Konrad pointed out with genetic data was that it’s discontinuous (NOT a continuous manifold). I don’t use rnaseq or UMAP in my own research so I am truly a non expert on this subject! I’m really happy to hear from people who use it about their use cases.
Also not an expert, just delving into ML and clustering algorithms. This episode really made me think about how much this and other algorithms just show you what you want to see if you play enough with the hyperparameters.